New Microbiome Pipeline Improves Forensic Body Fluid Identification
Researchers from the DQBM Kümmerli Lab and an international team have published a new study in Applied and Environmental Microbiology, presenting a standardized pipeline for identifying body fluids from complex forensic samples using microbiome analysis and machine learning.
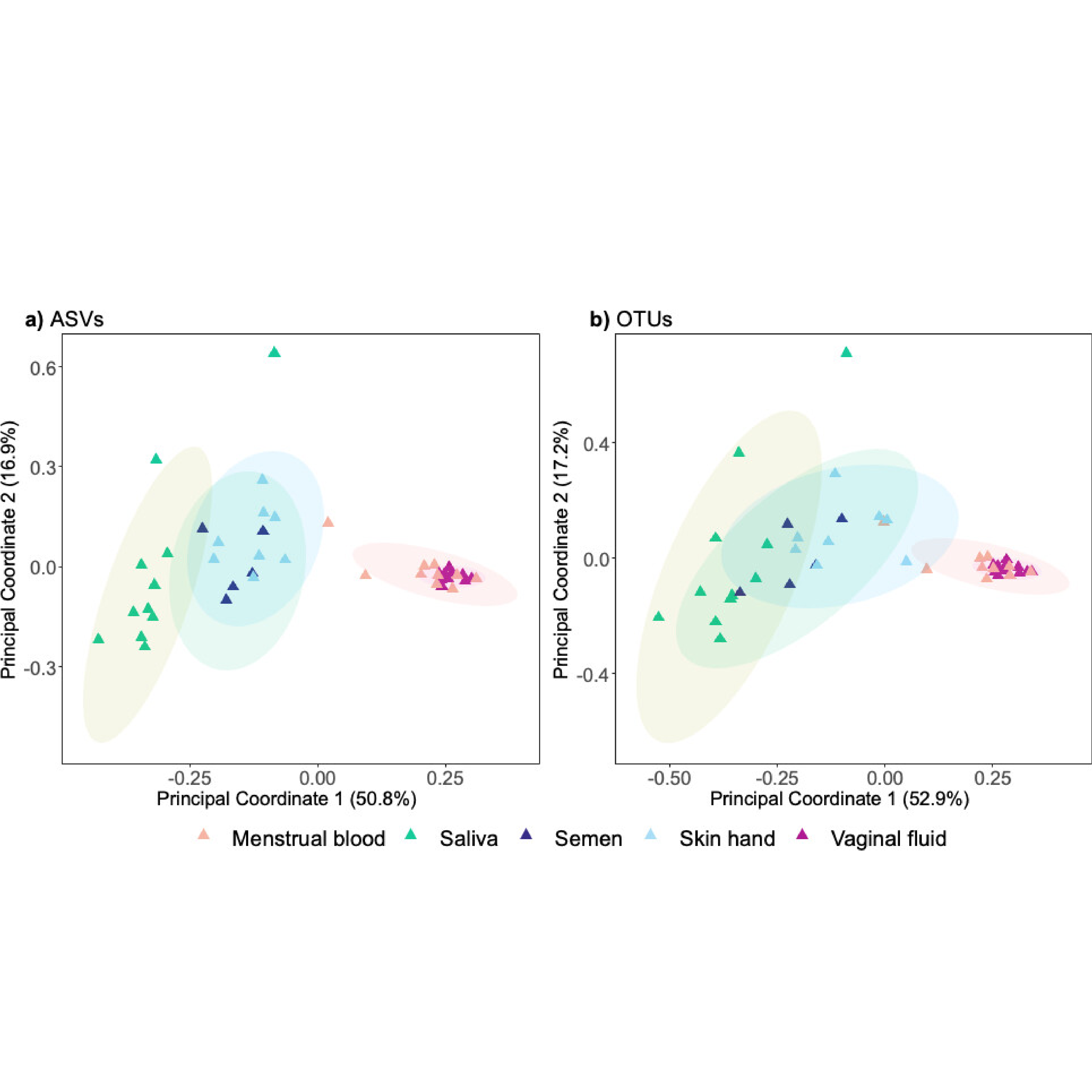
Current forensic practices often struggle to reliably identify biological fluids in complex or mixed samples. Addressing this, the research team developed standardized bioinformatics workflows and machine learning protocols to robustly classify body fluids such as saliva, semen, vaginal fluid, urine, and skin samples based on microbial composition. Their method successfully utilized operational taxonomic units (OTUs) derived from various microbial gene regions, training a random forest classifier to accurately predict sample origin.
Validation on realistic forensic samples, including mixtures and substrates like clothing, demonstrated high classification accuracy, achieving an overall reliability (F1 score) of 0.89. Additionally, the pipeline effectively identified sexually shared microbiota, providing new avenues for reconstructing criminal events in sexual assault cases.
This research significantly advances the application of microbiome analysis in forensic science, potentially transforming the accuracy and interpretative power of forensic evidence in criminal investigations.
Publication Link: https://doi.org/10.1128/aem.01871-24