Predicting Gene Editing Outcomes with Deep Learning
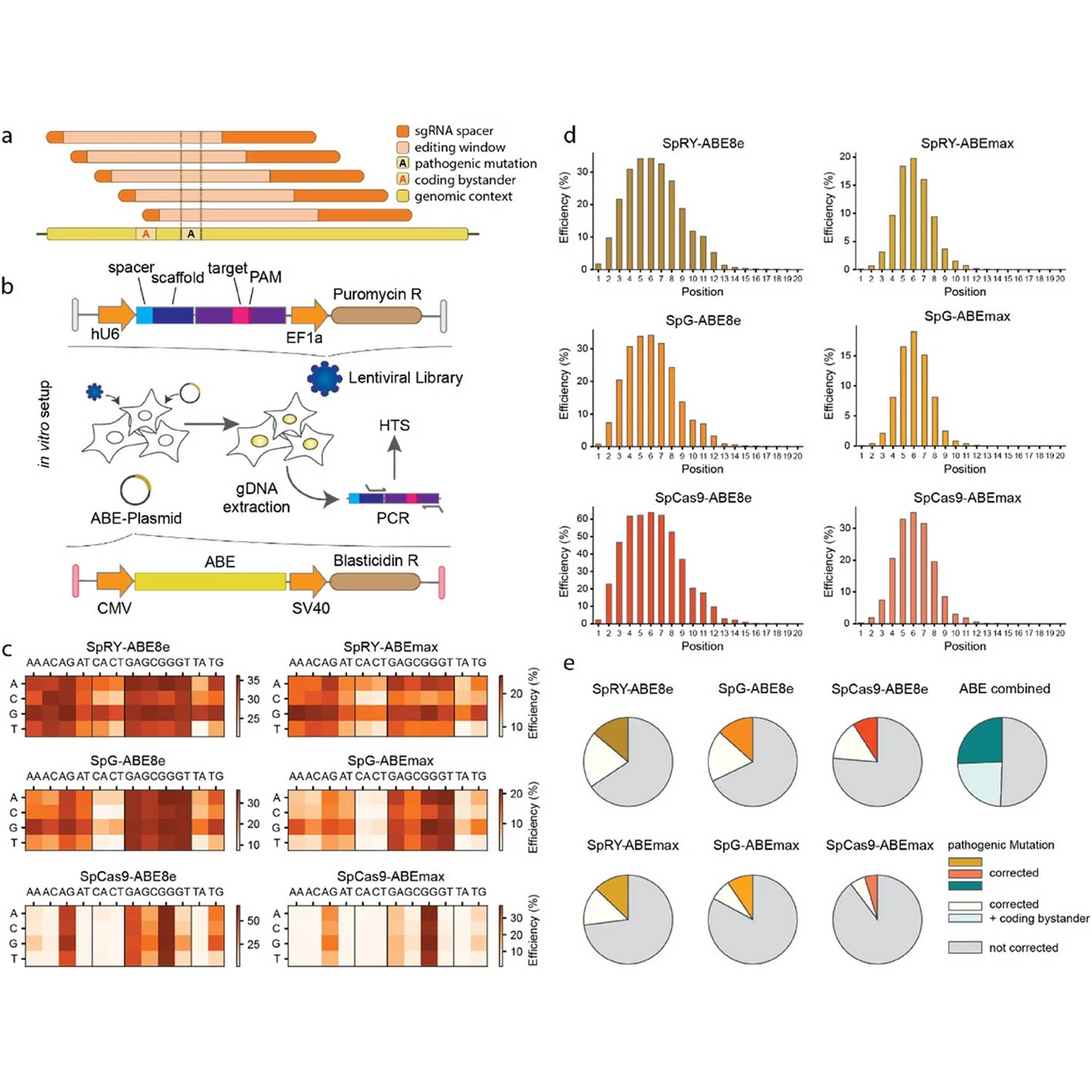
A collaborative study involving the Krauthammer Lab at the Department of Quantitative Biomedicine (DQBM), University of Zurich, has developed BEDICT2.0—a deep learning model that predicts the efficiency of adenine base editing (ABE) across diverse biological contexts. Published in Genome Biology on April 25, 2025, the research benchmarks the model using large-scale ABE screens targeting over 2,000 pathogenic mutations.
The team performed systematic ABE screens in human cells and in the murine liver, using multiple ABE variants delivered by plasmids, mRNA-lipid nanoparticles, and AAV vectors. Findings highlight that editing efficiencies differ by delivery method, with mRNA-based in vitro models better predicting in vivo outcomes.
BEDICT2.0 showed superior or comparable performance to existing models and was particularly effective when trained on datasets simulating physiological expression levels. This advancement supports more accurate in silico design of CRISPR therapies and enhances translational genome editing efforts.