Strain Identity Crucial for Bacterial Community Functioning
New research published in The ISME Journal, involving the Kümmerli Lab at DQBM, demonstrates that individual bacterial strain identities contribute more significantly to community functioning than previously assumed strain interactions.
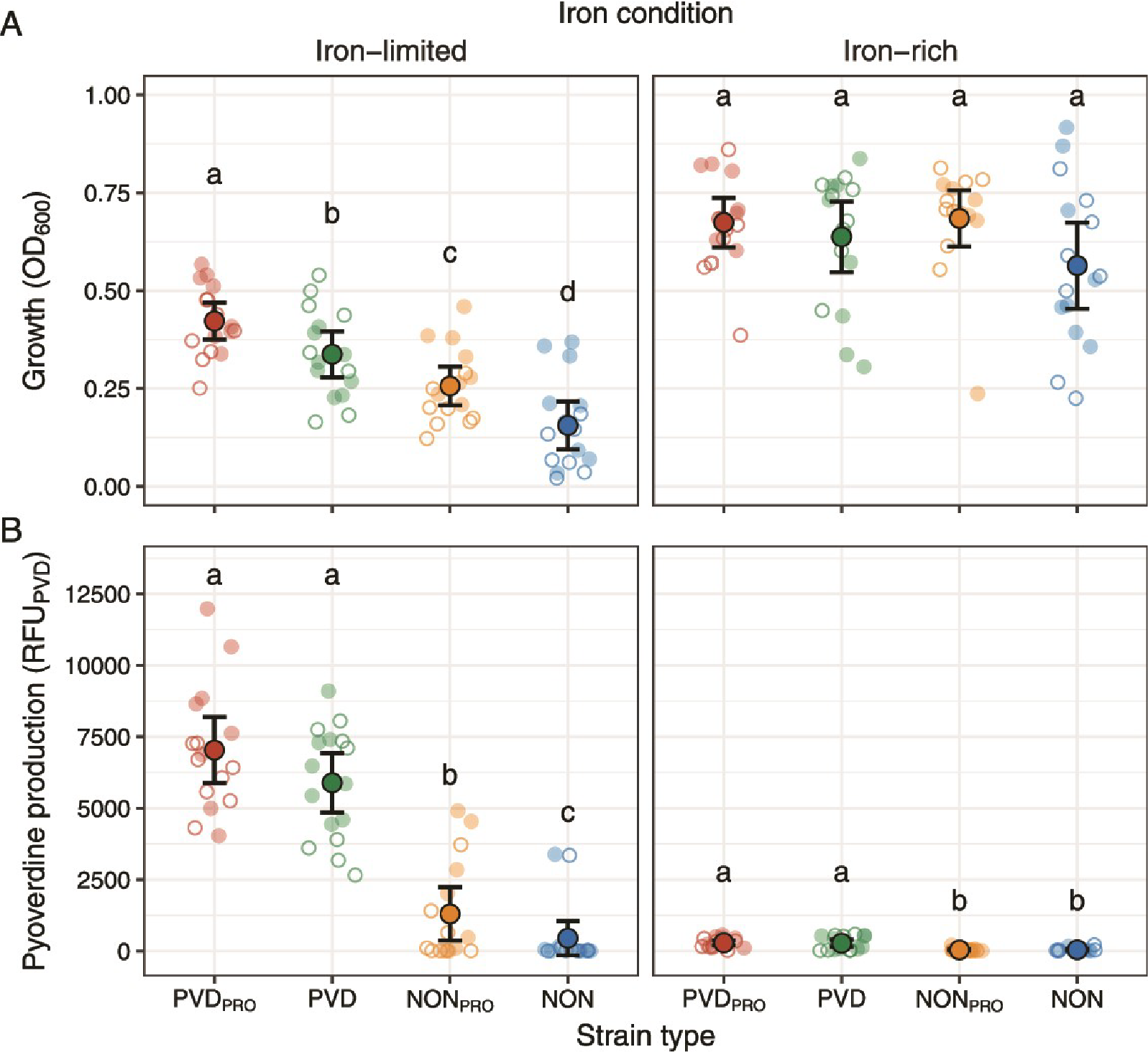
Using 64 strains of Pseudomonas bacteria isolated from diverse environmental settings, the researchers assessed the importance of strain identities and their interactions in determining two critical microbial functions: overall productivity and iron-scavenging siderophore production. Combining experimental and computational approaches, they found that while strain interactions—especially through secreted molecules like siderophores—were impactful, the individual characteristics of each strain predominantly dictated community behavior and productivity.
This result emphasizes that microbiome engineering strategies aimed at enhancing beneficial microbial interactions must account for inherent differences among bacterial strains. The research highlights a nuanced understanding of bacterial communities, informing approaches in biotechnology, environmental management, and human health.