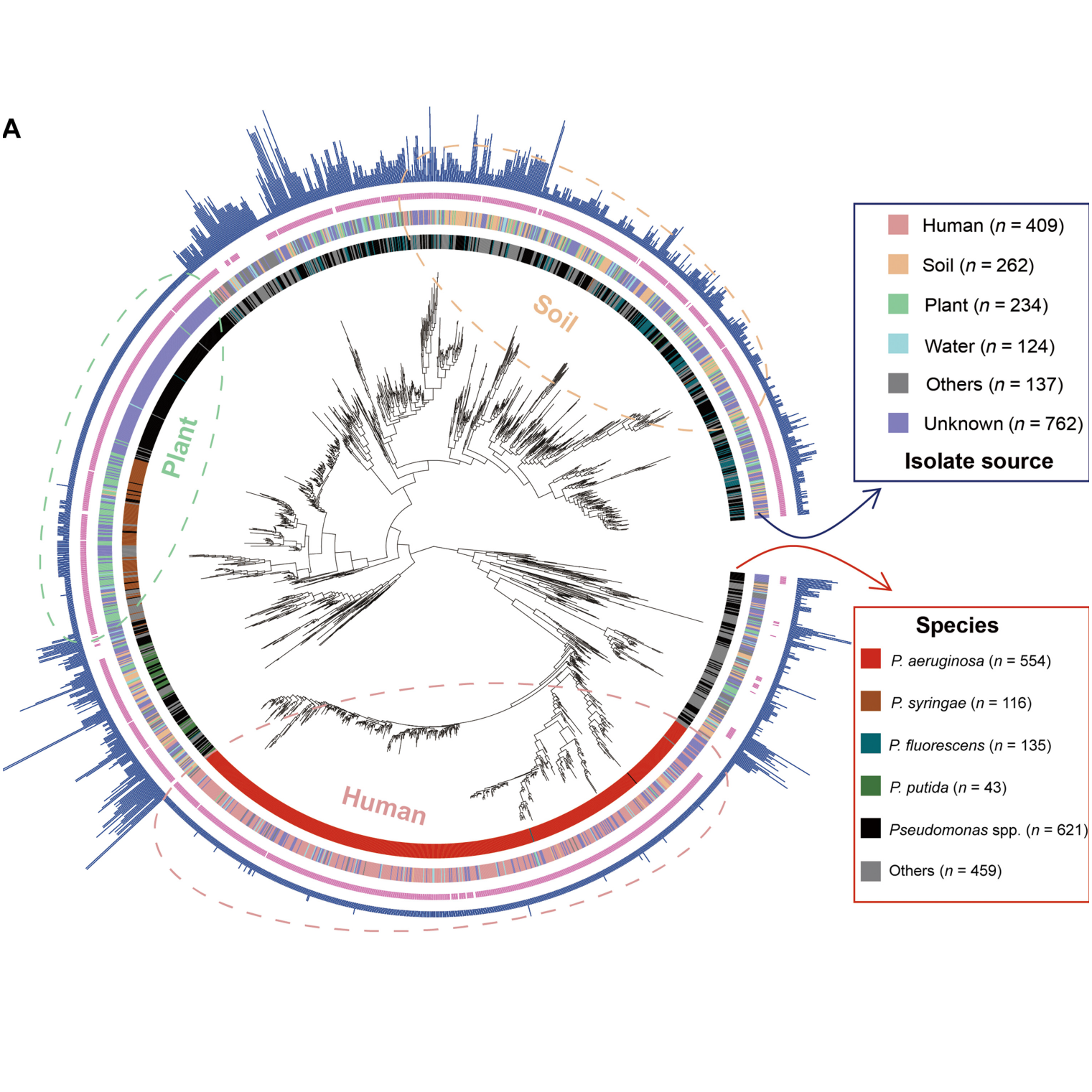
Coevolution Algorithm Reveals Bacterial Iron Interaction Networks
In a new study published in Science Advances, researchers from the DQBM Kümmerli Lab, together with international collaborators, developed an innovative computational approach to predict bacterial interactions through iron-scavenging molecules known as siderophores.
Iron is essential yet scarce, prompting bacteria to produce siderophores to scavenge iron from their environment. The team analyzed 1,928 genomes of Pseudomonas bacteria, developing a novel coevolution algorithm to identify precise matches between siderophore-producing genes and corresponding receptor genes. Their findings showed distinct bacterial interaction networks based on habitat and lifestyle: complex and highly interconnected in environmental, non-pathogenic bacteria, but simpler and fragmented in pathogenic strains.
These results suggest a selective pressure for exclusive iron acquisition strategies among pathogens, potentially informing targeted interventions against harmful bacteria. This new sequence-based tool advances our ability to understand and manipulate microbial communities, opening pathways for precision microbiome management in health, agriculture, and biotechnology.
Publication Link: https://doi.org/10.1126/sciadv.adq5038