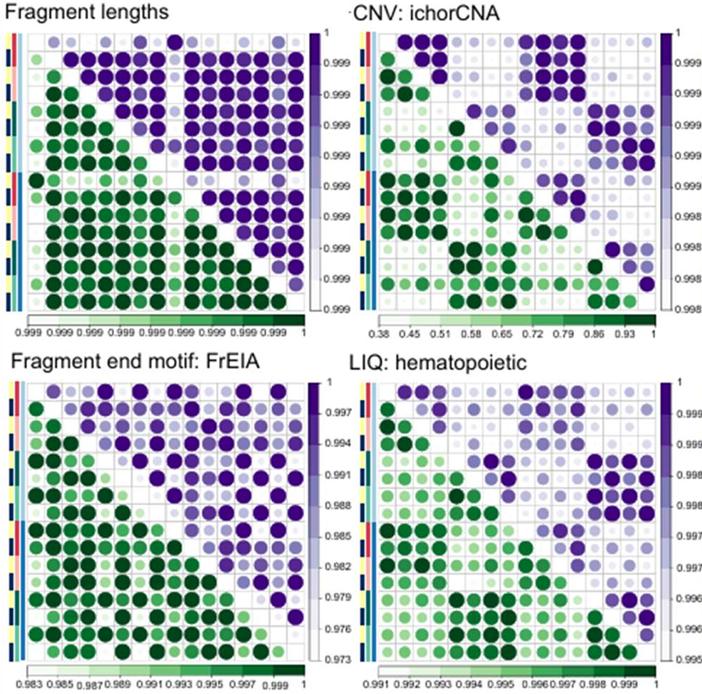
Researchers from the Krauthammer Lab at DQBM, with partners at Amsterdam UMC and the University of Manchester, assessed how trimming, alignment filtering, reference genomes, coverage, and in silico size selection influence key cfDNA features. Most genetic, fragmentomic, and epigenetic measures remained stable across 16 preprocessing settings; strict alignment filtering and hg38 modestly improved some copy number and nucleosome metrics.
They provide clear guidance: fragment lengths and end motifs work at 0.1x coverage, copy number calls need about 1x, and nucleosome footprints are highly coverage sensitive. Size selection boosts end motif classification. The team also releases cfDNA Flow, a modular Snakemake workflow for reproducible analyses. Published in GigaScience.